COSMOSIL RNA Series
RNA-based therapeutics represent a powerful new tool for gene therapy. In RNA production, impurities such as nucleotide triphosphates, abortive transcripts, and DNA templates can affect downstream applications by triggering immune responses. Gel-based methods of purification, such as preparative denaturing polyacrylamide gel electrophoresis (PAGE) and agarose gels, are limited by poor resolution of RNA size and potential chemical modifications from reagents such as formaldehyde. To date, high performance liquid chromatography (HPLC) remains a staple method for purification of messenger RNA (mRNA) and RNA oligonucleotides. Presented here are two HPLC methods: a size exclusion-based method for the purification of long mRNA, and a reversed-phase method for the purification of oligonucleotides.
- Detects impurities not observable with electrophoresis.
- Scale up to preparative separations.
- Separate long RNA strands from 100 to 5,000 bases.
- Two modes available: reversed-phase and size-exclusion.
References
>> Read our paper in Analytical Sciences on HPLC separation of RNA using RNA-RP1
Preparative separation of long-stranded ssRNA
Reversed-phase column: COSMOSIL RNA-RP1
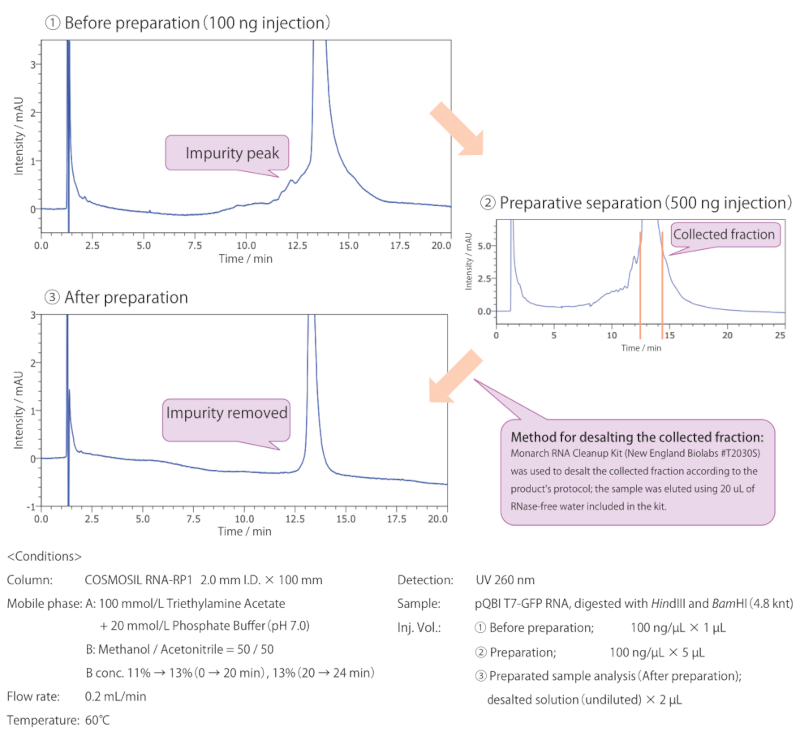
For this analysis, in vitro transcribed long-chain ssRNA (4.8 knt) precipitated using lithium chloride was used as the sample. When the long-chain ssRNA was analyzed using electrophoresis, the impurity was not detected (see page 6); however, using COSMOSIL RNA-RP1, a peak of an impurity thought to be of different length was detected (see chromatogram ① above). Using preparative separation by HPLC, the impurity peak was successfully removed (see chromatogram ③ above).
Two modes to choose from
The COSMOSIL RNA series is comprised of RNA-RP1, a reversed-phase column, and RNA-SEC-1000 and RNA-SEC-2000, size-exclusion columns. Chromatograms of these columns using the same ssRNA ladder are compared below.
Reversed-phase: COSMOSIL RNA-RP1
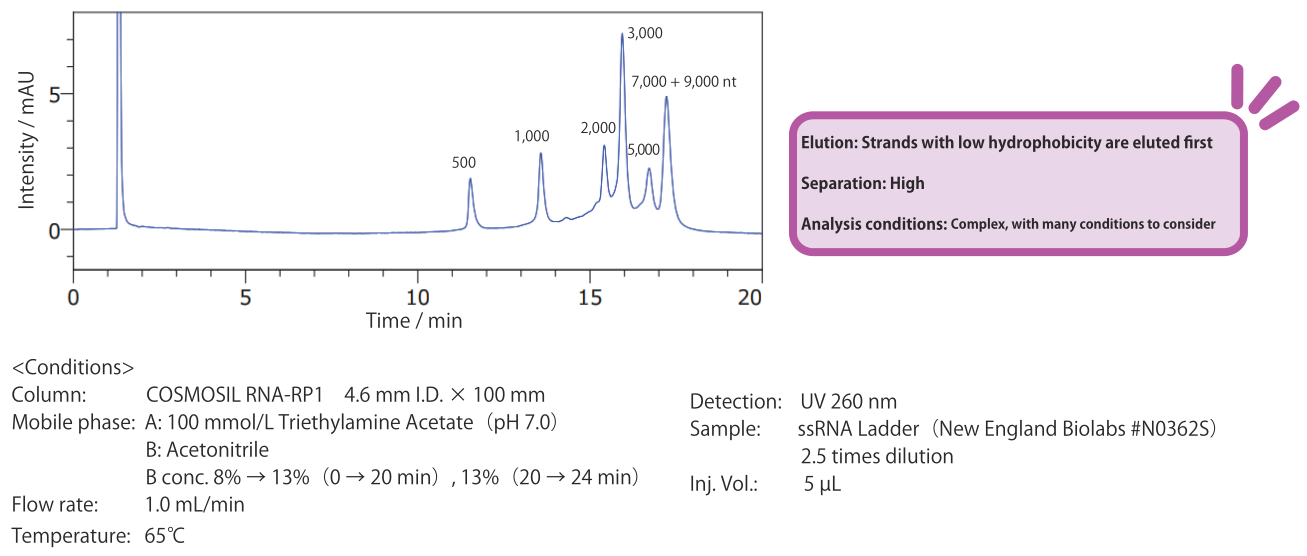
As the hydrophobicity of RNA strands generally increases with length, long strands are typically eluted after short strands. However, the hydrophobicity also differs by sequence, so it is not necessarily the case that strands are eluted in order of length. By optimizing the analysis conditions, it is possible to improve separation. For example, it is possible to adjust the mobile phase, temperature, and gradient slope. See the references on page 8 for details.
Size-exclusion: COSMOSIL RNA-SEC-1000, RNA-SEC-2000
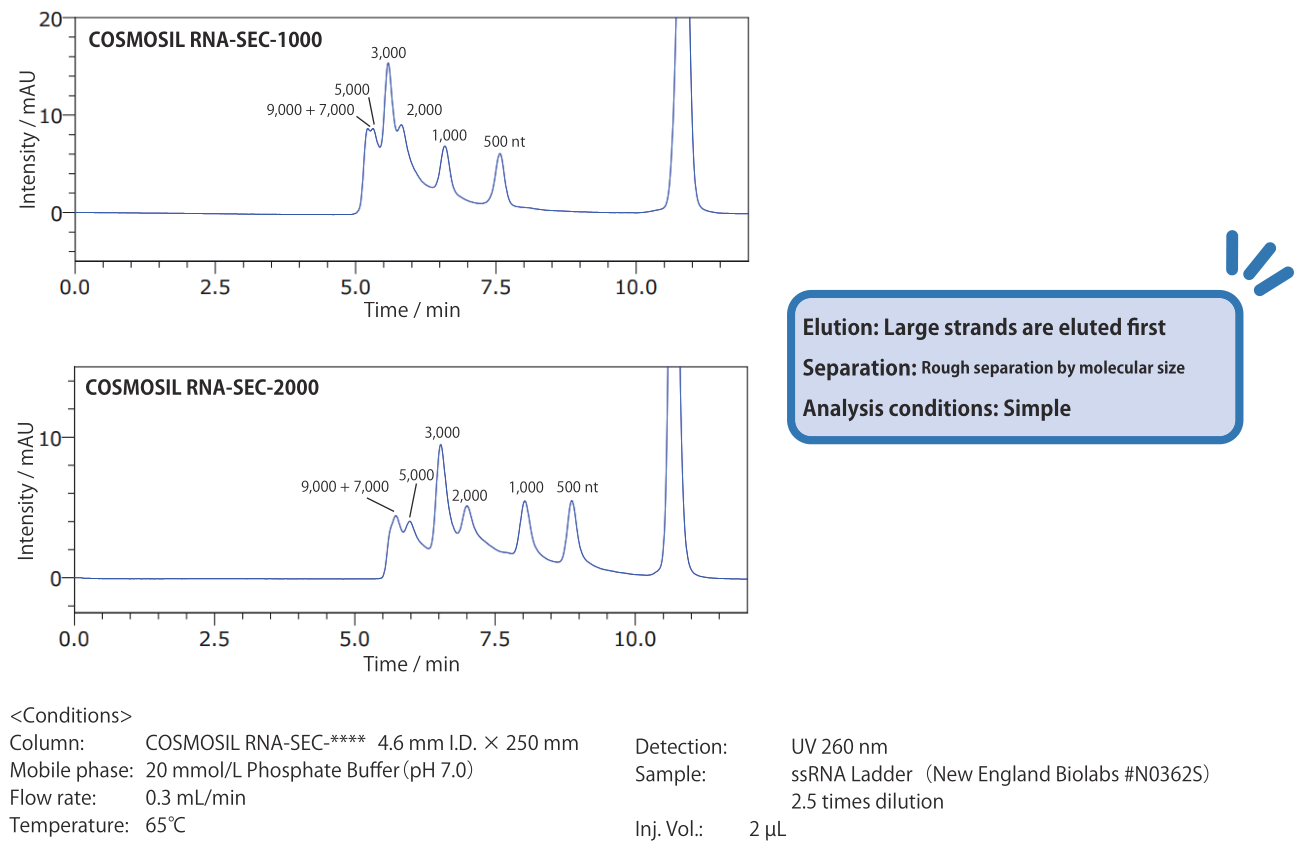
Compared to COSMOSIL RNA-SEC-1000, which has a pore size of 100 nm, RNA-SEC-2000, with a pore size of 200 nm, is suitable for analysis of large strands. Use different pore sizes depending on the strand sizes you need to separate.
Properties of packing material
Pore size
Long-strand nucleic acids have a very large molecular size, so they do not enter the pores of normal packing material, so the individual peaks do not separate. The super-wide pores allow the nucleic acids to enter, enabling much better separation.
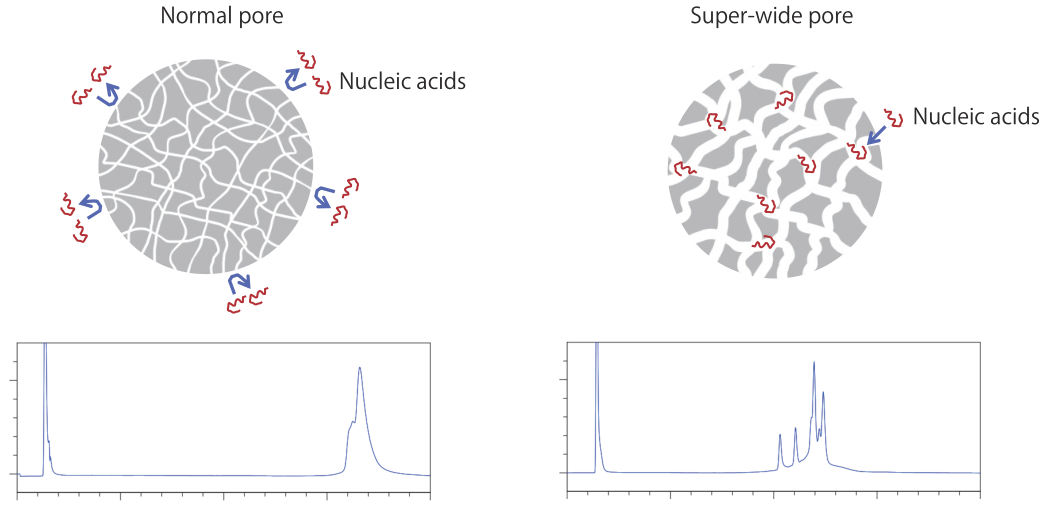
HPLC columns for general use typically have silica gel with an average pore size of 8 to 12 nm, whereas COSMOSIL RNA-RP1 uses silica gel with super-wide pores.
RNA-SEC-1000 and RNA-SEC-2000 use silica gel with 100 and 200 nm pores, respectively, so long-chain nucleic acids can enter the pores and be separated.
Target molecular sizes
Our recommendations for target molecular sizes for reversed-phase and size-exclusion columns are noted below.
* These figures only represent the range of general suitability; it does not indicate that any nucleic acids in the range can be separated.
- Suitable range for reversed-phase chromatography
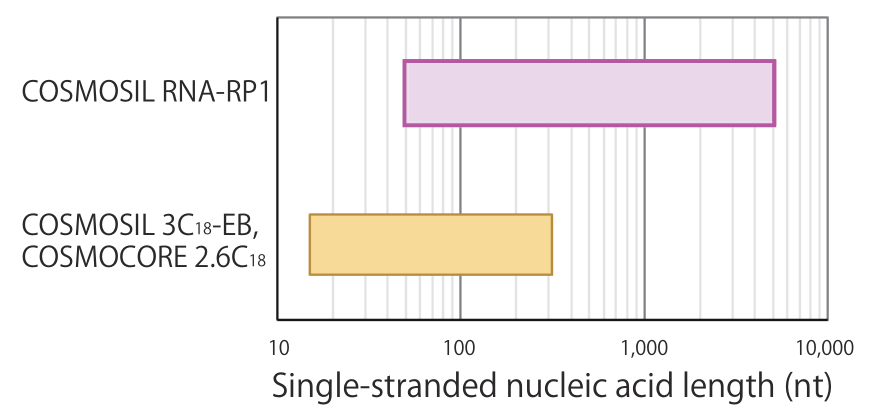
For mid-length to long-stranded nucleic acids above 100 nt, the super-wide pore silica gel in COSMOSIL RNA-RP1 is recommended. For oligonucleotides, columns with smaller pores, such as 3C18-EB (average pore size 12 nm) or COSMOCORE 2.6C18 (average pore size 9 nm) are recommended.
- Suitable range for size-exclusion chromatography
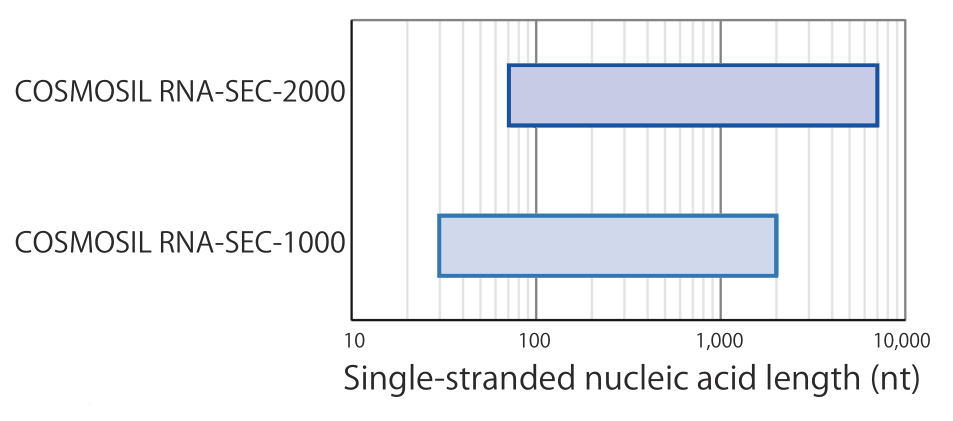
Using COSMOSIL RNA-SEC-1000, it is possible to separate smaller strands of less than 100 nt, up to long strands of thousands of nucleotides. Using RNA-SEC-2000, even longer strands of greater than 5,000 nt can be separated.
Analysis conditions
Analysis temperature
⬤ Single-strand nucleic acids
- Reversed-phase: COSMOSIL RNA-RP1
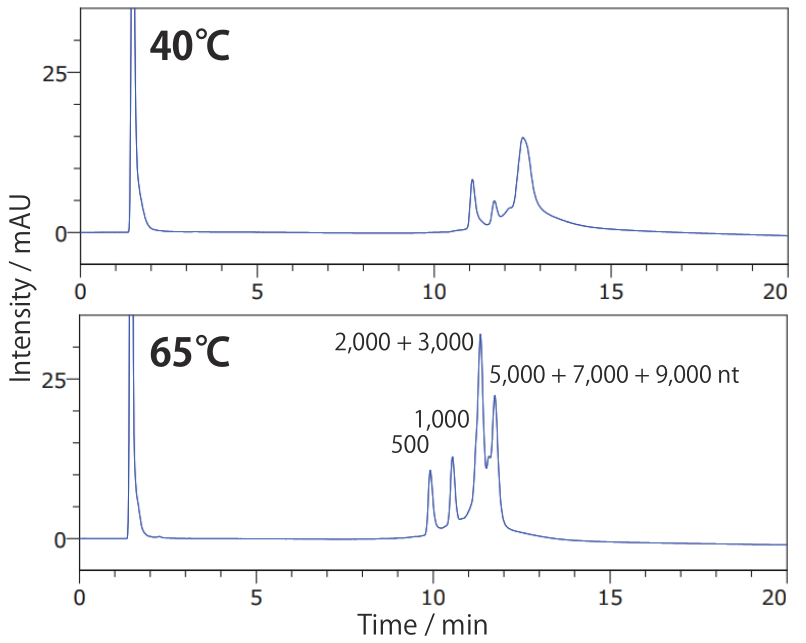
Conditions Column: COSMOSIL RNA-RP1 2.0 mm I.D. × 100 mm Mobile phase: A: 100 mmol/L Triethylamine Acetate(pH 7.0) B: Acetonitrile B conc. 7.5% → 17.5%(0 → 20 min), 17.5%(20 → 24 min) Flow rate: 0.2 mL/min Temperature: **°C Detection: UV 260 nm Sample: ssRNA Ladder(New England Biolabs #N0362S) 2.5 times dilution × 2 µL - Size-exclusion: COSMOSIL RNA-SEC-2000
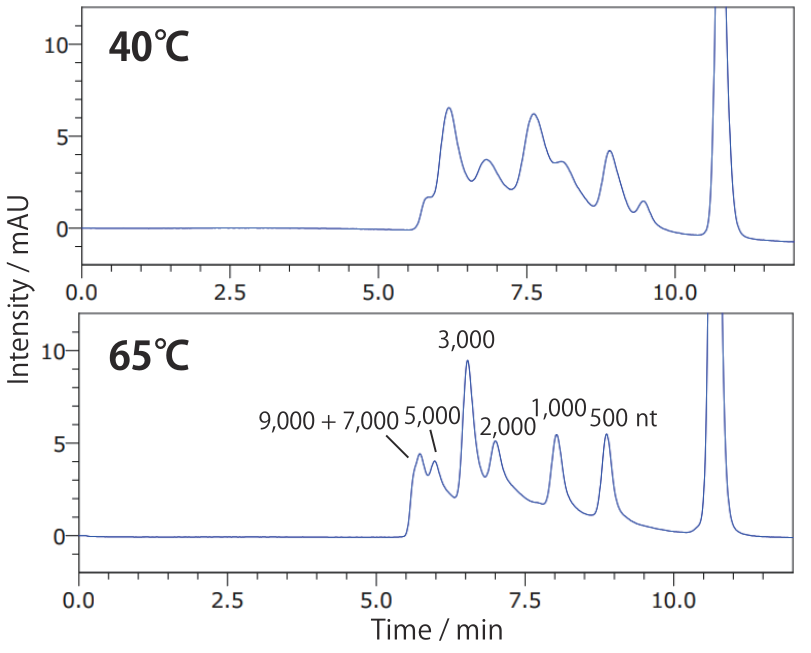
Conditions Column: COSMOSIL RNA-SEC-2000 4.6 mm I.D. × 250 mm Mobile phase: 20 mmol/L Phosphate Buffer (pH 7.0) Flow rate: 0.3 mL/min Temperature: **°C Detection: UV 260 nm Sample: ssRNA Ladder(New England Biolabs #N0362S) 2.5 times dilution × 2 µL
At low temperatures, single-strand nucleic acids tend to form higher-order structures, resulting in broad peaks. This maybe improved by analyzing at higher temperature.
⬤ Double-strand nucleic acids
- Reversed-phase: COSMOSIL RNA-RP1
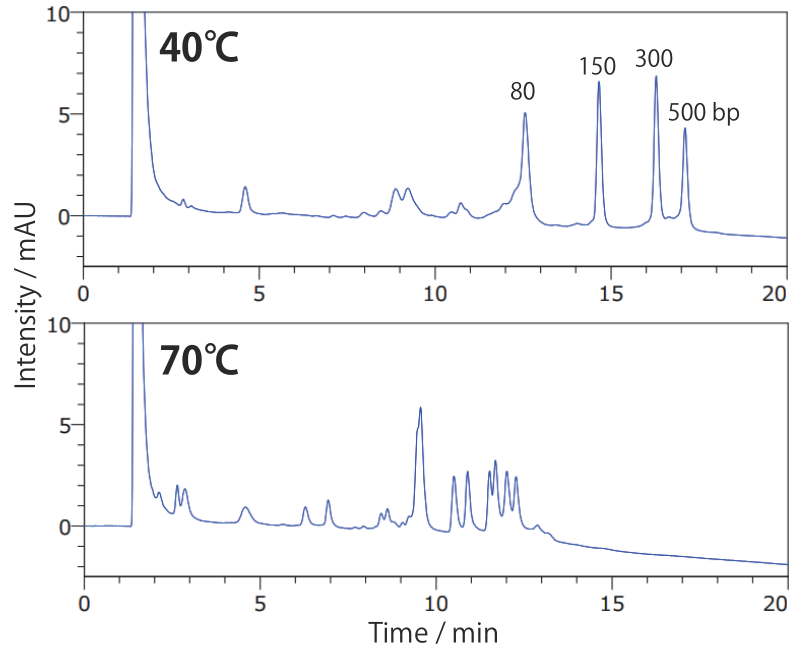
Conditions Column: COSMOSIL RNA-RP1 2.0 mm I.D. × 100 mm Mobile phase: A: 100 mmol/L Triethylamine Acetate(pH7.0) B: Acetonitrile B conc. 5% → 20%(0 → 20 min), 20%(20 → 24 min) Flow rate: 0.2 mL/min Temperature: **°C Detection: UV 260 nm Sample: dsRNA Ladder(New England Biolabs #N0363S) 2.5 times dilution × 2 µL
For analyzing double-strand nucleic acids in their nondenatured state, care is required when setting analysis temperature, as the strands tend to dissociate at high temperatures. Around the dissociation temperature, small differences in temperature can lead to very different separations. See our paper for details.
⬤ Mix of single- and double-strand nucleic acids
- Reversed-phase: COSMOSIL RNA-RP1
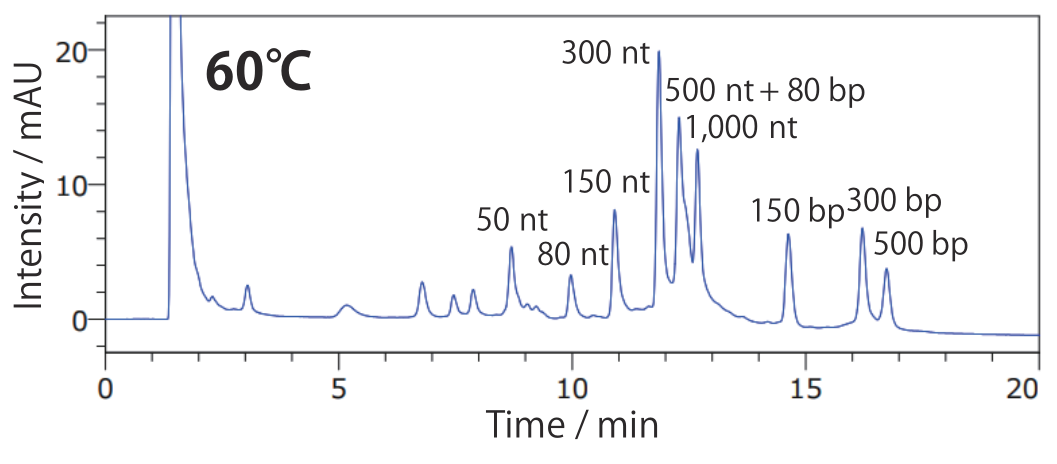
Conditions Column: COSMOSIL RNA-RP1 2.0 mm I.D. × 100 mm Mobile phase: A: 100 mmol/L Triethylamine Acetate(pH7.0) B: Acetonitrile B conc. 5% → 20%(0 → 20 min), 20%(20 → 24 min) Flow rate: 0.2 mL/min Temperature: 60°C Detection: UV 260 nm Sample: dsRNA Ladder(New England Biolabs #N0363S) 2.5 times dilution × 2 µL + Low Range ssRNA Ladder(New England Biolabs #N0364S)2.5 times dilution × 2 µL
By analyzing at a temperature at which single strands do not form higher-order structures and double strands do not dissociate, it is possible to simultaneously analyze both.
Considering loading capacity
- Size-exclusion: COSMOSIL RNA-SEC-1000
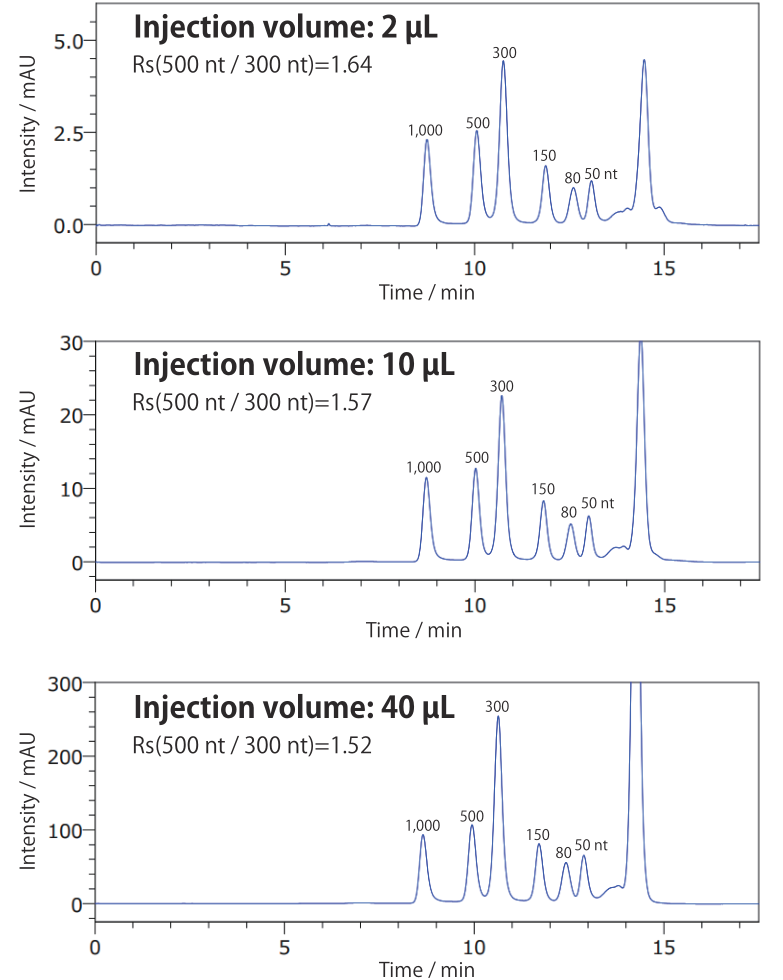
Conditions Column: COSMOSIL RNA-SEC-1000 7.5 mm I.D. × 300 mm Mobile phase: 20 mmol/L Phosphate Buffer (pH7.0) Flow rate: 1.0 mL/min Temperature: 65°C Detection: UV 260 nm Sample: Low Range ssRNA Ladder (New England Biolabs #N0364S)
Even injecting 40 uL of sample, a similar chromatogram to a 2 uL injection was obtained (vertical axis scale differs). Loading capacity differs by sample, so determine it experimentally using an analysis column before scaling to preparative size.
Application data
COSMOSIL RNA-RP1 (RNA ladder)
- Low Range ssRNA Ladder
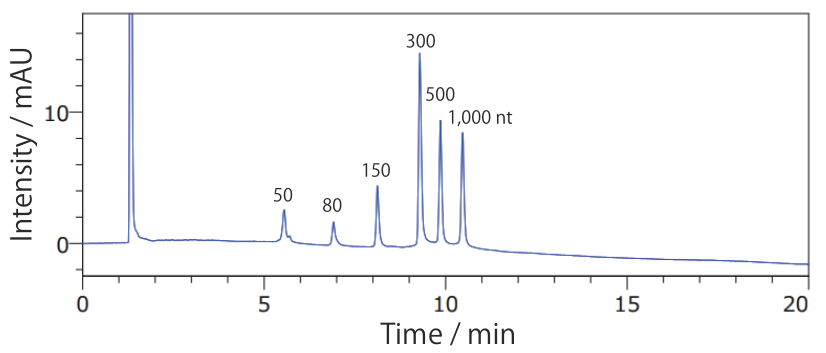
Conditions Column: COSMOSIL RNA-RP1 4.6 mm I.D. × 100 mm Mobile phase: A: 100 mmol/L Triethylamine Acetate(pH 7.0) B: Acetonitrile B conc. 5% → 20%(0 → 20 min), 20%(20 → 24 min) Flow rate: 1.0 mL/min Temperature: 65°C Detection: UV 260 nm Sample: Low range ssRNA Ladder(New England Biolabs #N0364S) 2.5 times dilution × 5 µL - ssRNA Ladder
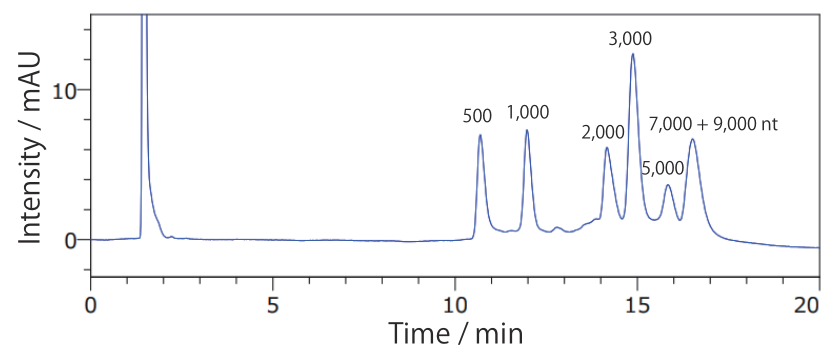
Conditions Column: COSMOSIL RNA-RP1 2.0 mm I.D. × 100 mm Mobile phase: A: 100 mmol/L Triethylamine Acetate(pH 7.0) B: Acetonitrile B conc. 7% → 10%(0 → 20 min), 10%(20 → 24 min) Flow rate: 0.2 mL/min Temperature: 65°C Detection: UV 260 nm Sample: ssRNA Ladder(New England Biolabs #N0362S) 2.5 times dilution × 2 µL
COSMOSIL RNA-RP1(DNA ladder)
- pBR322 / MspI digest
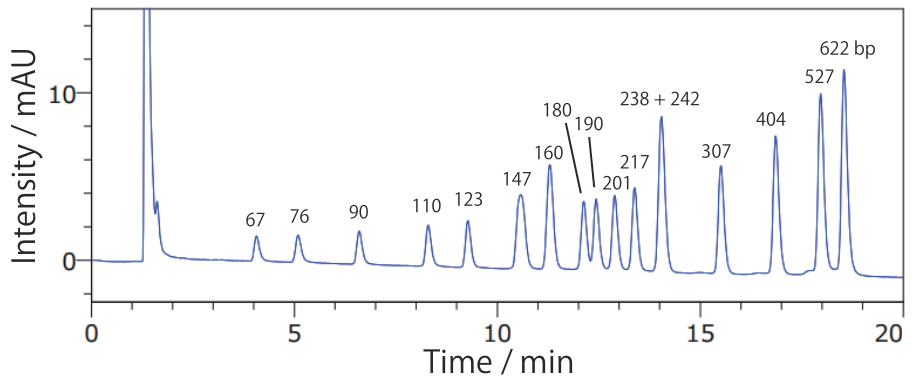
Conditions Column: COSMOSIL RNA-RP1 4.6 mm I.D. × 100 mm Mobile phase: A: 100 mmol/L Triethylamine Acetate(pH 7.0) B: Acetonitrile B conc. 10% → 17%(0 → 20 min), 17%(20 → 25 min) Flow rate: 1.0 mL/min Temperature: 40°C Detection: UV 260 nm Sample: pBR322 / MspI digest(Nippongene #317-01501) 200 ng/µL × 5 µL - 100 bp DNA Ladder
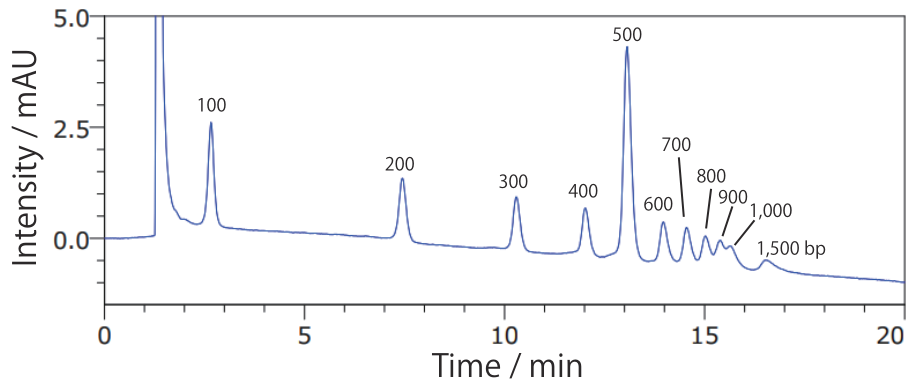
Conditions Column: COSMOSIL RNA-RP1 4.6 mm I.D. × 100 mm Mobile phase: A: 100 mmol/L Triethylamine acetate(pH 7.0) B: Acetonitrile B conc. 12% → 18%(0 → 20 min), 18%(20 → 25 min) Flow rate: 1.0 mL/min Temperature: 40°C Detection: UV 260 nm Sample: 100bp DNA Ladder(Takara Bio #3407A) 130 ng/µL × 5 µL
Fragments that are difficult to separate using agarose gel electrophoresis can be separated using COSMOSIL RNA-RP1.
COSMOSIL RNA-RP1 (separation of impurities)
- ssRNA synthesized by in vitro transcription
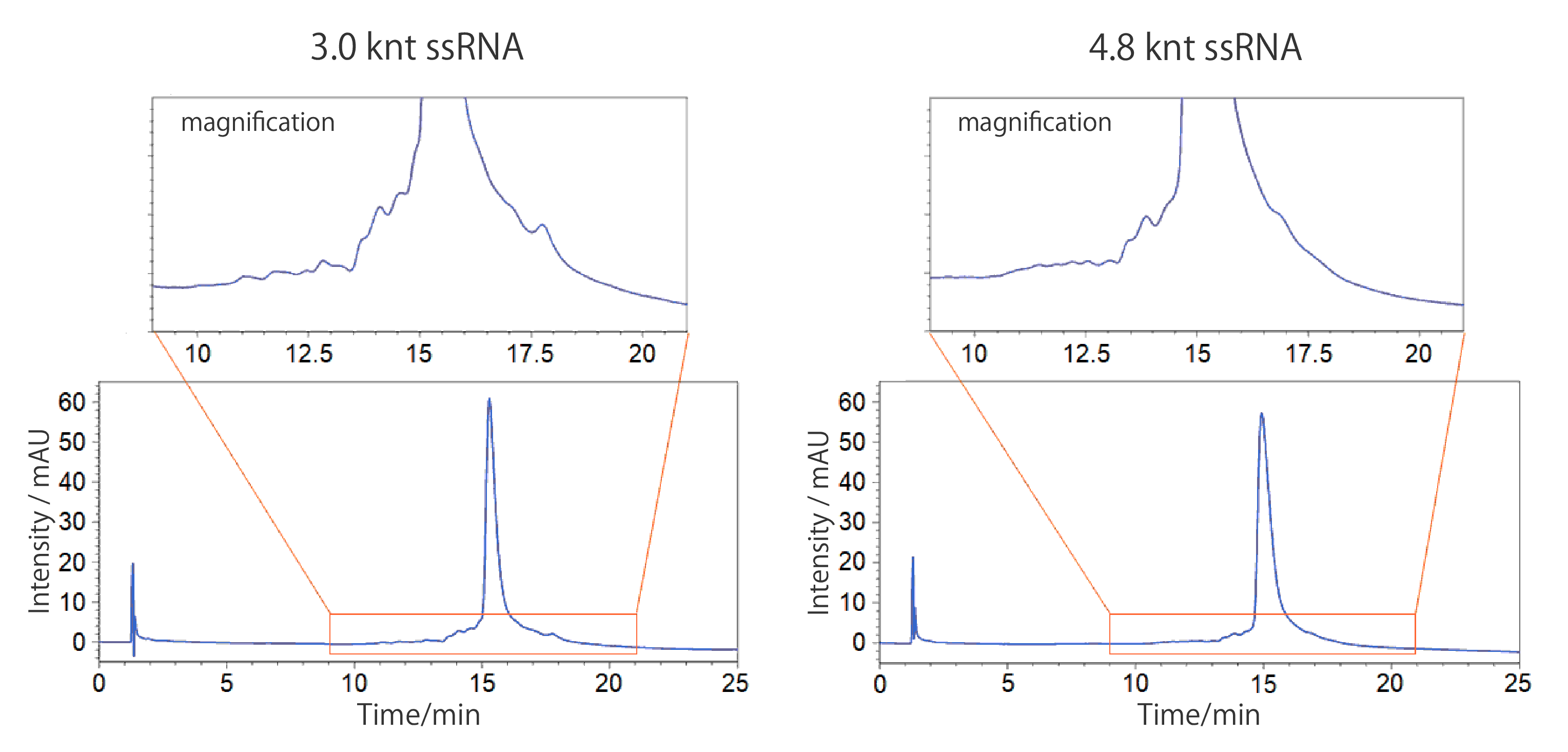
Conditions Column: COSMOSIL RNA-RP1 2.0 mm I.D. × 100 mm Mobile phase: A: 100 mmol/L Triethylamine Acetate + 20 mmol/L Phosphate Buffer (pH 7.0) B: Methanol / Acetonitrile = 50 / 50 B conc. 11% → 13%(0 → 20 min), 13%(20 → 24 min) Flow rate: 0.2 mL/min Temperature: 60°C Detection: UV 260 nm Sample: pQBI T7-GFP RNA, digested with BglII and EagI (3.0 knt), 100 ng/µL × 5 µL pQBI T7-GFP RNA, digested with HindIII and BamHI (4.8 knt), 100 ng/µL × 5 µL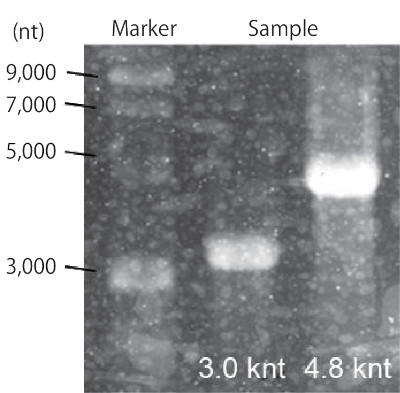
Upon analyzing long-chain ssRNA purified using lithium chloride precipitation, impurity peaks not detected with agarose gel electrophoresis were confirmed. This shows that it is possible to separate impurities in vitro transcribed ssRNA using reversed-phase HPLC.
COSMOSIL RNA-SEC-1000・RNA-SEC-2000(RNA ladder)
- Low Range ssRNA Ladder(65°C)
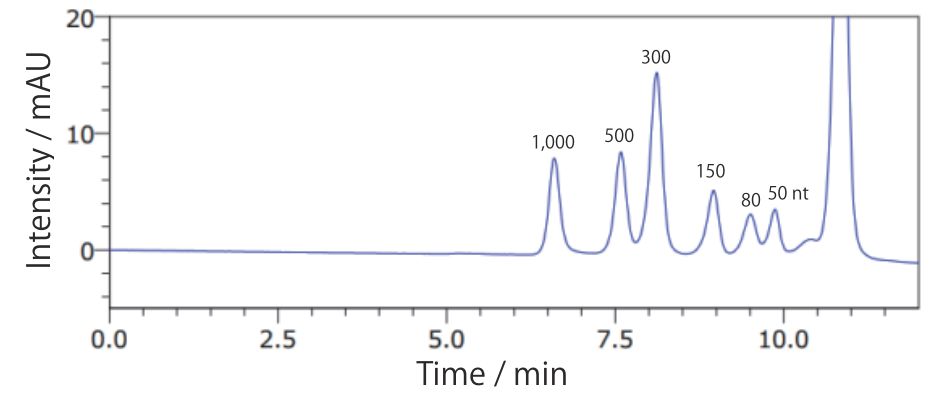
Conditions Column: COSMOSIL RNA-SEC-1000 4.6 mm I.D. × 250 mm Mobile phase: 20 mmol/L Phosphate Buffer(pH 7.0) Flow rate: 0.3 mL/min Temperature: 65°C Detection: UV 260 nm Sample: Low range ssRNA Ladder(New England Biolabs #N0364S) 2.5 times dilution × 2 µL - dsRNA Ladder(40°C)
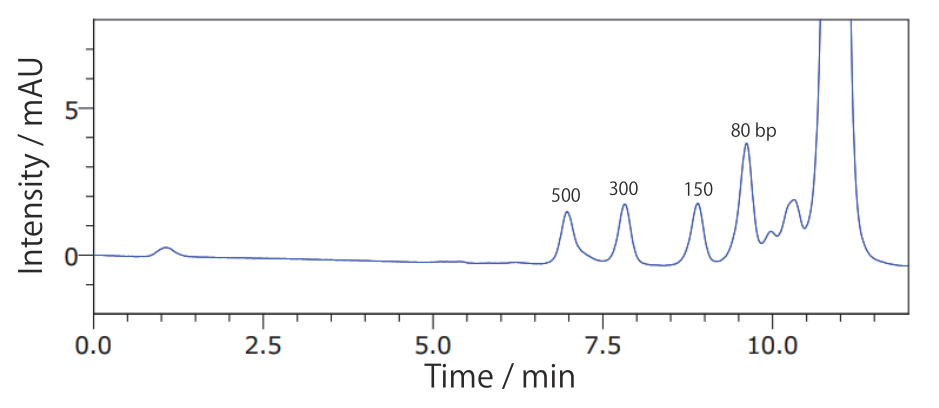
Conditions Column: COSMOSIL RNA-SEC-1000 4.6 mm I.D. × 250 mm Mobile phase: 20 mmol/L Phosphate Buffer(pH 7.0) Flow rate: 0.3 mL/min Temperature: 40℃ Detection: UV 260 nm Sample: dsRNA Ladder(New England Biolabs #N0363S) 2.5 times dilution × 2 µL - ΦX174 / HincII digest
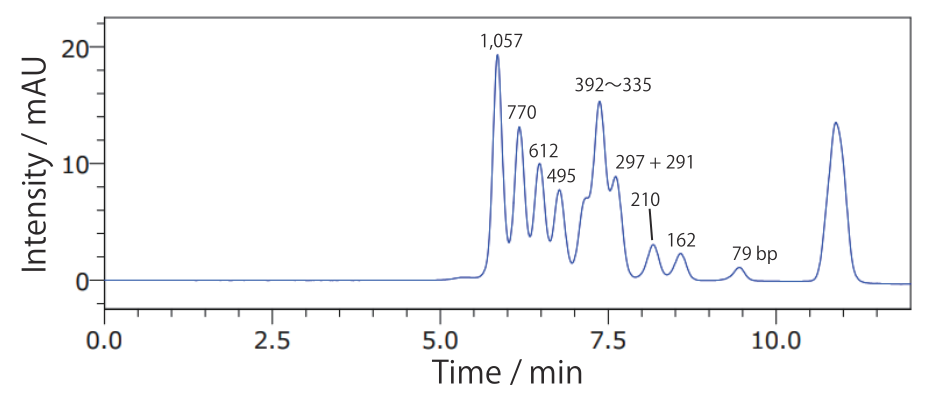
Conditions Column: COSMOSIL RNA-SEC-1000 4.6 mm I.D. × 250 mm Mobile phase: 20 mmol/L Phosphate Buffer(pH 7.0) Flow rate: 0.3 mL/min Temperature: 40°C Detection: UV 260 nm Sample: ΦX174 / HincII digest(Takara Bio #3406A) 200 ng/µL × 1.25 µL - 20 bp DNA Ladder
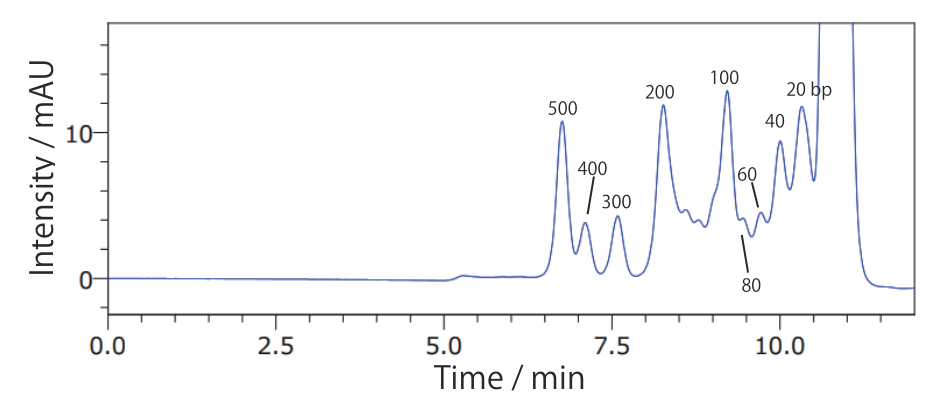
Conditions Column: COSMOSIL RNA-SEC-1000 4.6 mm I.D. × 250 mm Mobile phase: 20 mmol/L Phosphate Buffer(pH 7.0) Flow rate: 0.3 mL/min Temperature: 40°C Detection: UV 260 nm Sample: 20bp DNA Ladder(Takara Bio #3409A) 200 ng/µL × 2.5 µL - ΦX174 / HaeIII digest
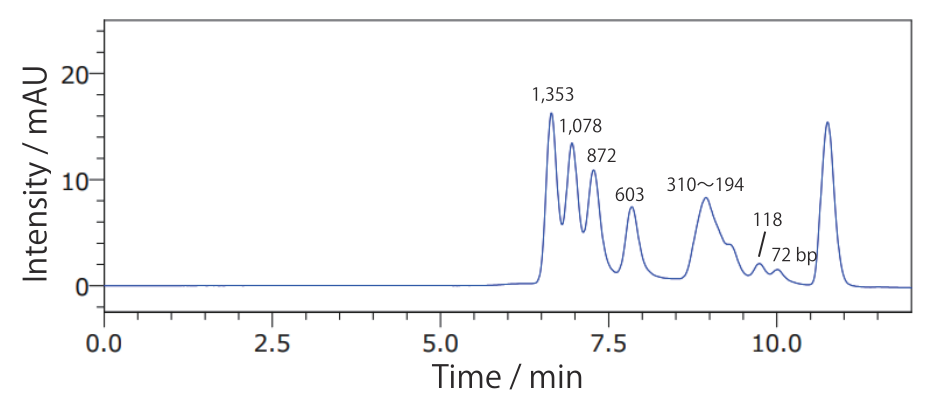
Conditions Column: COSMOSIL RNA-SEC-2000 4.6 mm I.D. × 250 mm Mobile phase: 20 mmol/L Phosphate Buffer(pH 7.0) Flow rate: 0.3 mL/min Temperature: 40°C Detection: UV 260 nm Sample: ΦX174 / HaeIII digest(Takara Bio #3405A) 200 ng/µL × 1.25 µL - 100 bp DNA Ladder
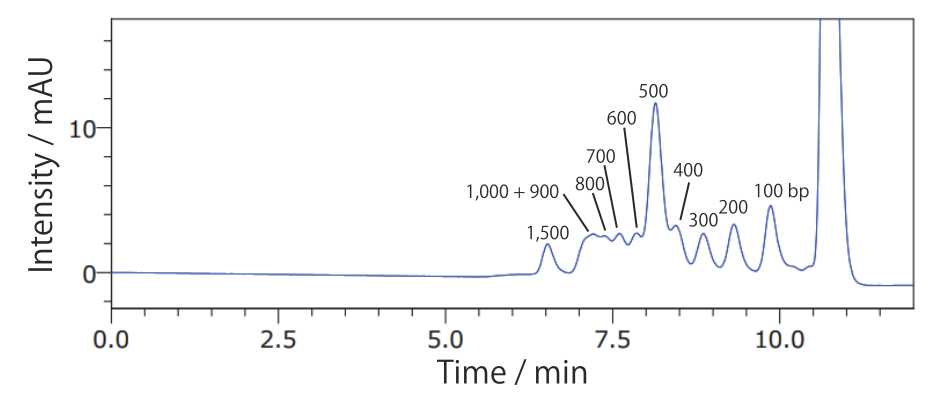
Conditions Column: COSMOSIL RNA-SEC-2000 4.6 mm I.D. × 250 mm Mobile phase: 20 mmol/L Phosphate Buffer(pH 7.0) Flow rate: 0.3 mL/min Temperature: 40°C Detection: UV 260 nm Sample: 100bp DNA Ladder(Takara Bio #3407A) 130 ng/µL × 4 µL
Product specifications
| Packing Material | RNA-RP1 | RNA-SEC-1000 | RNA-SEC-2000 |
|---|---|---|---|
| Separation mode | Reversed-phase | Size-exclusion | |
| Silica gel | Fully-porous high-purity spherical silica gel | ||
| Average particle size (µm) | 5 | Average pore size (nm, approx.) | — | 100 (1,000 Å) | 200 (2,000 Å) |
| Bonded phase | Octadecyl group | Hydrophilic group | |
| Suitable pH range | 2 – 7.5 | ||
| Maximum pressure (MPa) | 15 | ||
Ordering information
Other sizes may be available. Please contact us.